Cancer evolves. Those two words may sound strange together. Sure, birds evolve. Bacteria evolve. But cancer? The trouble arises from the fact that cancers, unlike birds and bacteria, are not free-living organisms. They start out as cells inside a person’s body and stay there, until they’re either wiped out or the person dies.*
Yet the same forces that drive the evolution of free-living organisms can also drive cancer cells to become more aggressive and dangerous. Evolution becomes our inner foe if mutations disable a cell’s self-restraint. The cell multiplies. Sometimes a new mutation arises in its descendants. If the mutations allow the cancer to grow faster, the cells carrying it will take over the population of cancerous cells. Natural selection and other processes that drive evolution on the outside start driving it on the inside.
Like so many other scientists, researchers who study cancer evolution have jumped on new technology for sequencing genomes on the cheap. They’re now starting to publish fine-grained histories of the disease, tracking individual mutations as they arise and spread. Nature has just published a fine example of this new research. I particularly appreciated the informative pictures they came up with to accompany the paper, one of which I’ve included here. You can click on the picture for a bigger version. And below the picture, I’ll explain what it means.
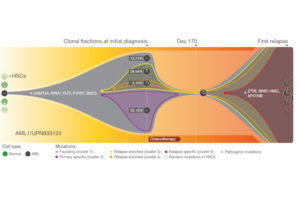
In the new paper, Li Ding and colleagues at Washington University describe a study they carried out on eight people suffering from acute myeloid leukemia (AML), a disease of the immune system. In people with AML, stem cells in the bone marrow that would normally turn into white blood cells instead become cancerous. Treatments include bone marrow transplants and chemotherapy. Unfortunately, AML has a nasty way of bouncing back from chemotherapy, and the drugs become useless to stop it. As a result, a lot of people who seem at first to be in remission eventually die of the cancer.
The Washington University scientists reconstructed the history of the cancer in each patients by sequencing genomes from a number of cells. To determine the normal, original genome, they sequenced DNA from a healthy skin cell. They then sequenced genomes from cancer cells taken from the patients when they were first diagnosed. And then they looked at genomes of cancer cells that emerged after the patients relapsed. From this survey, they came up with a catalog of new mutations that emerged over the course of the cancer. They could then go back into the blood samples and estimate what fraction of the cancer cells had a given mutation at a given point in time.
This figure illustrates the sad chronicle of one particular woman they studied. When she was in her late 50s, she suddenly came down with a sore throat and began to bruise easily. A bone marrow biopsy confirmed she has AML. She got chemotherapy, and then a stem cell transplant. Although she seemed to go into complete remission, the cancer returned 11 months after her diagnosis. The chemotherapy drugs that had previously been so effective now could not stop the cancer. Other drugs failed, too. Two years after her diagnosis, she died.
On the left of the figure, the cancer begins. A single stem cell mutated and became the founder of the cancerous lineage. we start with normal cells. (The cell is dark, and the grey dot marks its original mutation. HSC stands for hematopoietic stem cells).
The cancer cells grew in number, and as they did, they accumulated a lot of mutations, some of which are listed in the figure next to the star. All of these mutations, one after the other, took over the entire population of cells–a signature of natural selection. When the woman went to her doctor, however, the cancer had diversified into a number of different lineage, each carrying additional, distinctive mutations. Over half of the cells belonged to a lineage marked here in purple, known as cluster 2. Cluster 3, marked in yellow, was made up cells with a separate set mutations. And from within Cluster 3 emerged yet another lineage–Cluster 4, marked in orange. The dots in each circle show the sets of mutations that accumulated in each cluster.
The chemotherapy knocked down all the clusters of cancer cells to such low numbers that doctors couldn’t find them any more. But they were still there. And when exposed to chemotherapy drugs, the most successful cluster was not the one that had been most successful back when the cancer was diagnosed. It was the relatively rare Cluster 4. Apparently, it had mutations that made it better able to withstand the chemotherapy drugs. Some its descendants later picked up new mutations, which enabled them to reproduce quickly and take over the cancer population, as they resisted new chemotherapy drugs as well.
“The AML genome in an individual patient is clearly a “moving target,'” the scientists right conclude. “Eradication of the founding clone and all of its subclones will be required to achieve cures.” Easier said than done, of course. The parallels between this research and studies on antibiotic resistance in bacteria are sobering. But at least now we’re starting to see what kind of evolutionary challenge we’re really up against.
( *For one very cool exception to this rule, consider the case of Tasmanian devil facial tumors, which travel from devil to devil. They evolve too, though.)
Originally published January 12, 2012. Copyright 2012 Carl Zimmer.